Highlights
- •Arc is a multifunctional hub protein.
- •Arc is flexible, modular and capable of reversible self-oligomerization.
- •Arc localization and stability are regulated by post-translational modifications.
- •Arc protein complexes are implicated in the genetics of human cognition.
- •Arc is a master organizer of long-term synaptic plasticity.
Abstract
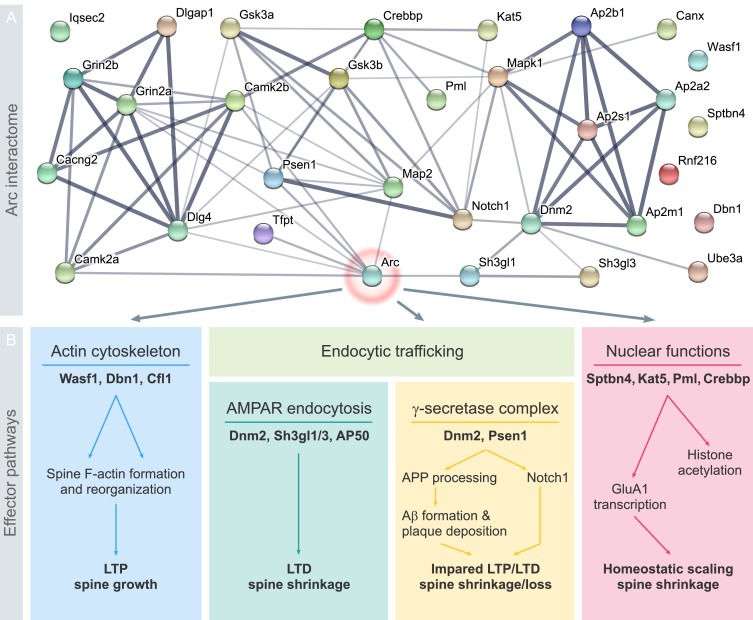
Mammalian excitatory synapses express diverse types of synaptic plasticity. A major challenge in neuroscience is to understand how a neuron utilizes different types of plasticity to sculpt brain development, function, and behavior. Neuronal activity-induced expression of the immediate early protein, Arc, is critical for long-term potentiation and depression of synaptic transmission, homeostatic synaptic scaling, and adaptive functions such as long-term memory formation. However, the molecular basis of Arc protein function as a regulator of synaptic plasticity and cognition remains a puzzle. Recent work on the biophysical and structural properties of Arc, its protein-protein interactions and post-translational modifications have shed light on the issue. Here, we present Arc protein as a flexible, multifunctional and interactive hub. Arc interacts with specific effector proteins in neuronal compartments (dendritic spines, nuclear domains) to bidirectionally regulate synaptic strength by distinct molecular mechanisms. Arc stability, subcellular localization, and interactions are dictated by synaptic activity and post-translational modification of Arc. This functional versatility and context-dependent signaling supports a view of Arc as a highly specialized master organizer of long-term synaptic plasticity, critical for information storage and cognition.